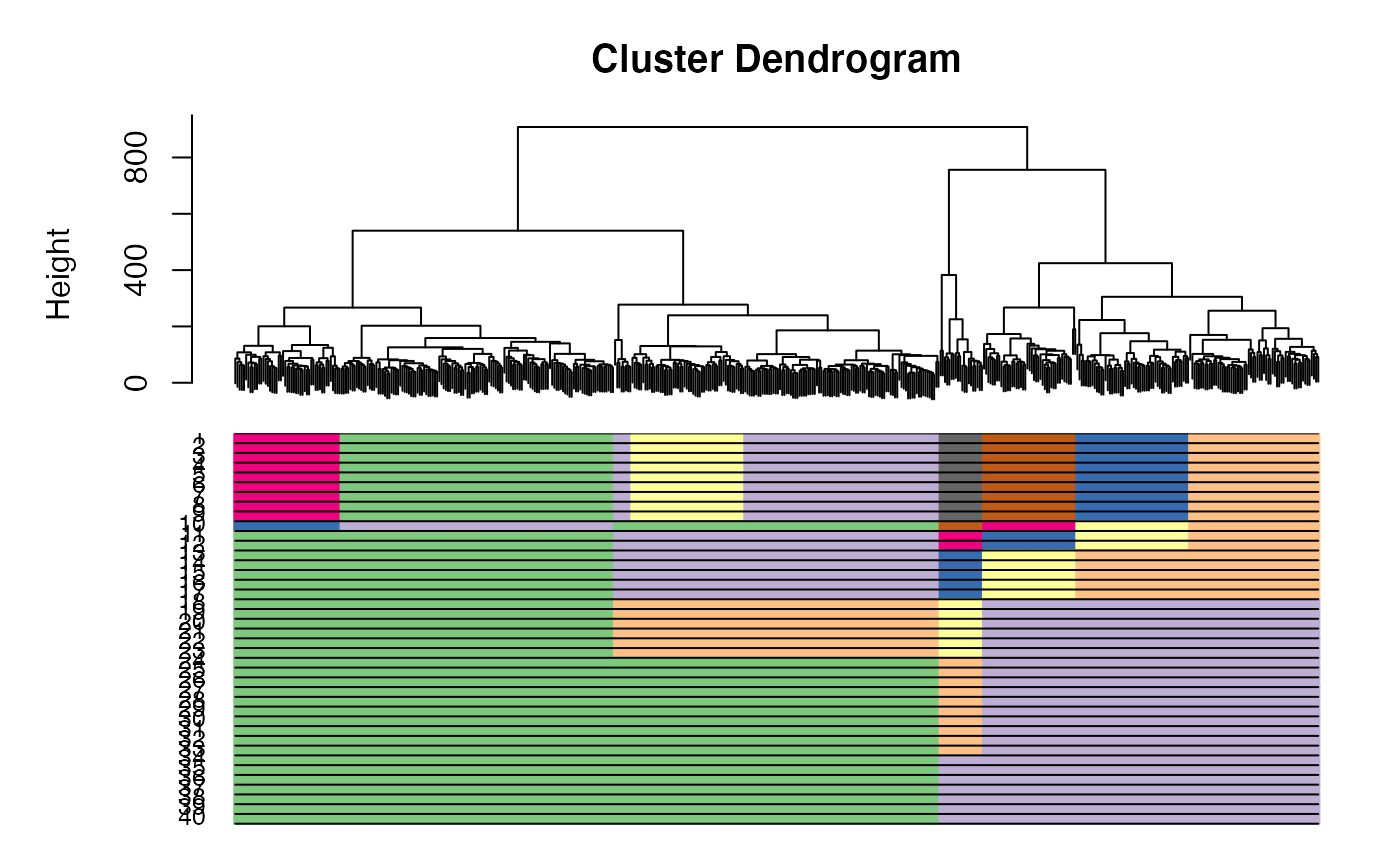
This function plots CORE and all clustering results underneath
plot_CORE(original.tree, list_clusters = NULL, color_branch = NULL)
Arguments
| original.tree | the original dendrogram before clustering |
|---|---|
| list_clusters | a list containing clustering results for each of the |
| color_branch | is a vector containing user-specified colors (the number of unique colors should be equal or larger than the number of clusters). This parameter allows better selection of colors for the display. |
Value
a plot with clustering bars underneath the tree
Examples
day5 <- day_5_cardio_cell_sample cellnames <- colnames(day5$dat5_counts) cluster <-day5$dat5_clusters cellnames <-data.frame('Cluster'=cluster, 'cellBarcodes' = cellnames) mixedpop2 <-new_summarized_scGPS_object(ExpressionMatrix = day5$dat5_counts, GeneMetadata = day5$dat5geneInfo, CellMetadata = cellnames) CORE_cluster <- CORE_clustering(mixedpop2, remove_outlier = c(0))#>#>#>#>#>#>#>#>#>#>#>#>#>#>plot_CORE(CORE_cluster$tree, CORE_cluster$Cluster)